Pharmacokinetic/Pharmacodynamic Modeling of Schedule-Dependent Interaction between Docetaxel and Cabozantinib in Human Prostate Cancer Xenograft Models
Wenjun Chen, Rong Chen, Jian Li, Yu Fu, Liang Yang, Hong Su, Ye Yao, Liang Li, Tianyan Zhou*, Wei Lu*, J Pharmacol Exp Ther 364:13–25, January 2018
Abstract
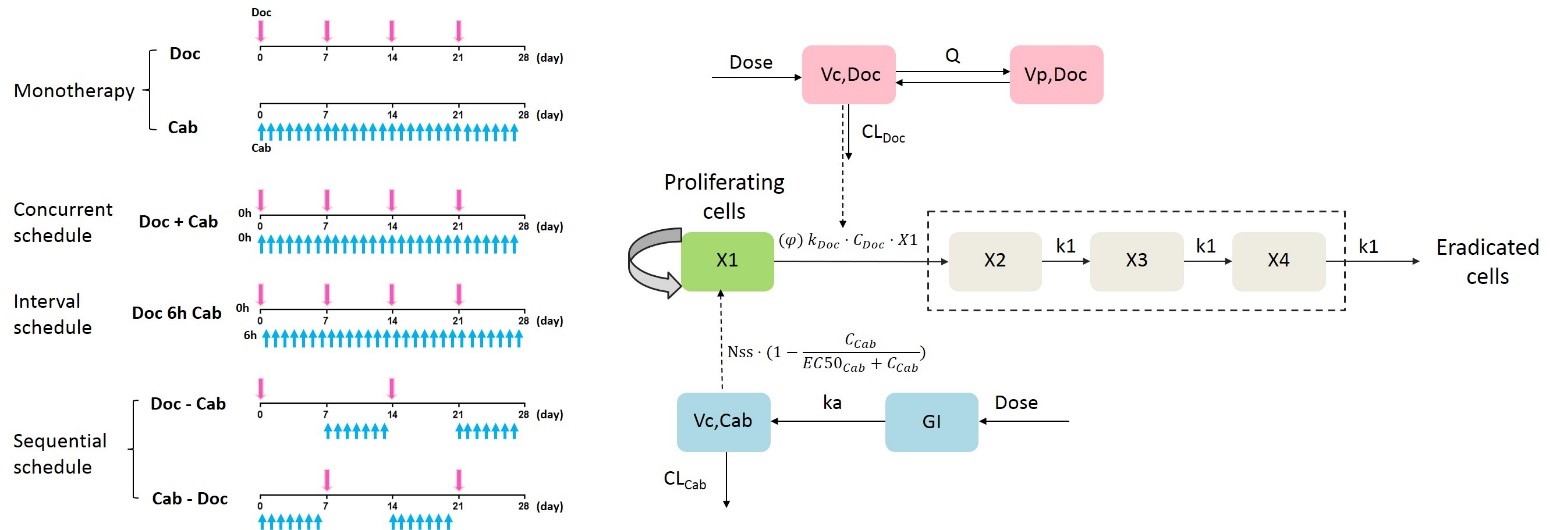
In this work, a semi-mechanistic pharmacokinetic/pharmacodynamic (PK/PD) model to quantitatively describe the antitumor activity of docetaxel (Doc) and cabozantinib (Cab) under monotherapy, concurrent therapy, interval therapy and different sequential therapy in mouse xenograft models of castration-resistant prostate cancer (CRPC) was developed and evaluated. Pharmacokinetics of Doc and Cab when administered separately and simultaneously were investigated in nude mice, and pharmacodynamic study was conducted in tumor-bearing mice treated with different dosing schedules. The PK interaction between Doc and Cab was expressed by adding the effect of Cab on the clearance of Doc in PK model. And the PD interaction between the two drugs was demonstrated by the developed PK/PD model through combination index “φ”. Our results showed that the concurrent therapy and Doc followed by Cab (Doc ~ Cab) sequential therapy exhibited better tumor inhibitory efficacy than monotherapy. The Cab followed by Doc (Cab ~ Doc) sequential schedule was less effective than monotherapy, and the interval therapy did not enhance the anti-tumor efficacy compared with the concurrent therapy. Parameter “φ” estimated from the PK/PD model quantitatively characterized the action between Doc and Cab. There was no significant PD interaction between Doc and Cab in both concurrent schedule and interval schedule, while the effect of the two drugs in “Doc ~ Cab” and “Cab ~ Doc” sequential schedule was synergistic and antagonistic, respectively. The proposed model properly described the anti-tumor effects of Doc and Cab under different treatment schedules, and could be used for dose optimization through model-based simulation.
Graphical Abstract:
http://URL of PubMed:https://www.ncbi.nlm.nih.gov/pubmed/29084815